This guide shows you how to organize an information retrieval shared task or experiment with ir_datasets in TIRA. Please contact the TIRA admins or comment below this thread if you want to organize a Shared Task using TIRA and have any questions or problems, we are happy to help.
Preparation
Please update the FROM clause of your docker image from FROM webis/tira-ir-datasets-starter:0.0.47 to FROM webis/tira-ir-datasets-starter:0.0.54. Version 0.0.47 had a minor issue so that it was not possible to render search engine result pages if no relevance judgments (qrels) were available that is fixed in version 0.0.54.
For the rest of the tutorial, we assume you have the following:
- A local docker image
<YOUR-DOCKER-IMAGE>containing your ir_datasets installation (please ensure it starts withFROM webis/tira-ir-datasets-starter:0.0.54as described above) - You have tested
<YOUR-DOCKER-IMAGE>locally withtira-run - You have a TIRA account and you are in two groups:
tira_org_ir-lab-sose23andtira_vm_ir-lab-sose-2023-<YOUR-TEAM>.
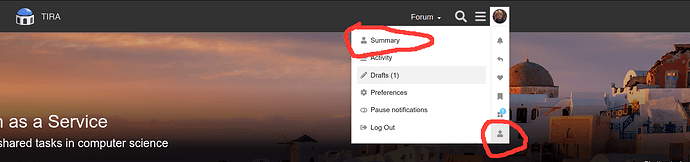
To check that you are in the correct group, go to your profile summary (click on your account → Profile → Summary):
On your profile, click “Expand” to show your groups, there you should find that you are in the two groups:
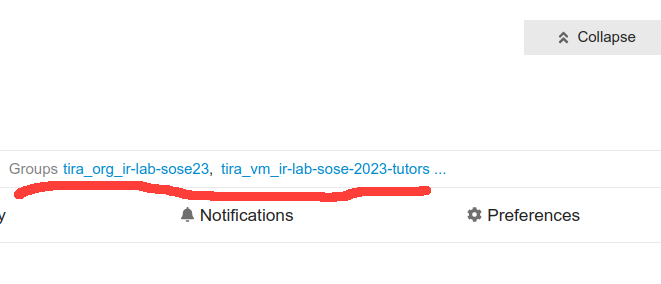
If you are not in the groups, please drop me a message.
Import your Dataset
Step 1: Navigate to https://www.tira.io/task/ir-lab-jena-leipzig-sose-2023
Step 2: Click on “Import Existing Dataset”
Step 3: Upload your docker image
In case you already have uploaded your image to docker hub, you can skip this step.
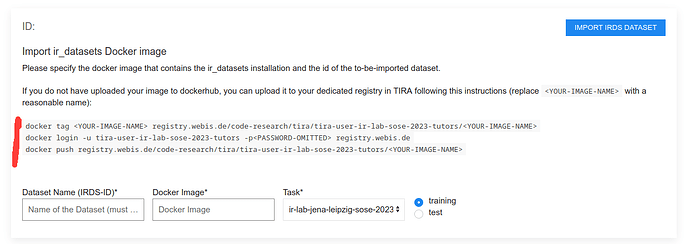
If you do not have a docker hub account, you can follow the instructions to upload your image to your dedicated docker registry in TIRA:
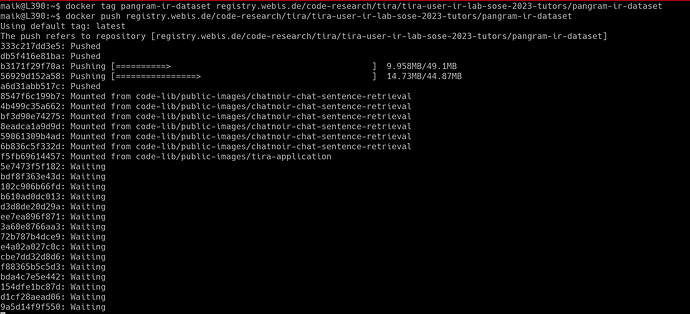
For instance, to upload the pangram docker image from the tutorial, the commands might look like this (you have to replace tira-user-ir-lab-sose-2023-tutors with your group):
docker tag pangram-ir-dataset registry.webis.de/code-research/tira/tira-user-ir-lab-sose-2023-tutors/pangram-ir-dataset
docker login ...
docker push registry.webis.de/code-research/tira/tira-user-ir-lab-sose-2023-tutors/pangram-ir-dataset
which should look like this:
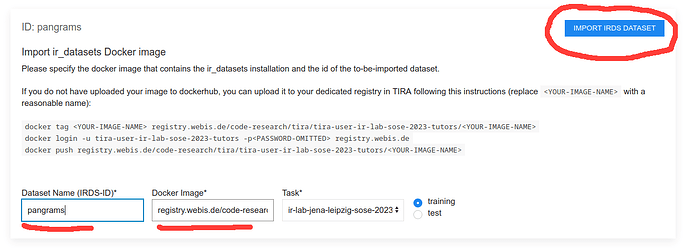
Step 4: Fill out the import form and upload
First, Specify the name of your dataset, and the docker image in the formular, then click on “Import IRDS Dataset”. The formular might look like this:
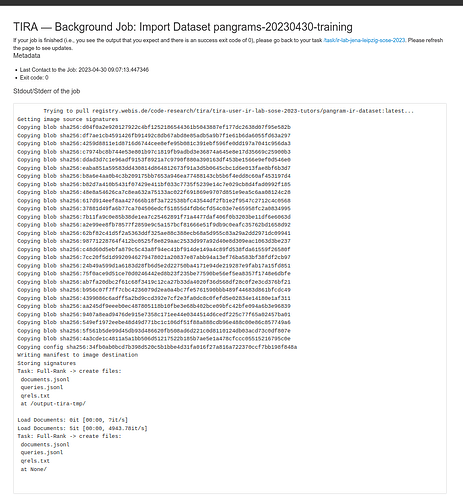
After clicking on “Import IRDS Dataset” you are redirected so that you see the progress of the import.
Please refresh this page regularly to see the progress (auto refresh is on the todo list :)), as soon as the output is as expected (i.e., the return code of the import indicates success, i.e., 0 and the stdout/stderr is as you would expect it), you are done ![]()
A valid import might look like this:
If you spot an error, you can delete your dataset and create it new from scratch.
Double-check that everything is correct
After you have imported your dataset as specified above, you can check that everything is as expected.
Step 1: Navigate to https://www.tira.io/task/ir-lab-jena-leipzig-sose-2023
Step 2: Select your dataset and go to its settings
Step 3: Download the data and verify it on your system
After clicking on the settings for your datasets, scroll to “Export Dataset” and click the link “Download input for Systems”:
This will download the dataset to your system so that you can verify it.
If you spot an error, you can delete your dataset and create it new from scratch.